Staff
- Professor
Satoshi TAKAHASHI
- Assistant Professor
Yuji ITOH
Outline
- Dynamics of protein folding revealed by single molecule fluorescence spectroscopy
- Functional dynamics of a tumor suppressor p53 by single molecule fluorescence microscopy
- Development of new strategy of protein design
Single molecule and ensemble observation on protein folding dynamics
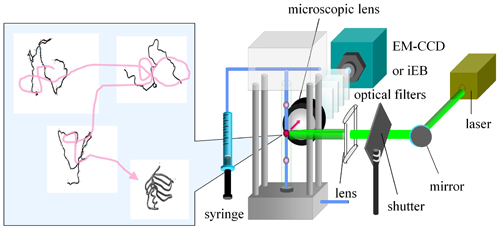
Proteins fold from the fully unfolded state to the functional native state based on the information coded in the amino acid sequences. To understand the molecular mechanism of protein folding, we investigate the dynamic processes of protein folding based on single molecule fluorescence spectroscopy. Furthermore, to reveal the functions of a tumor suppressor p53, we track the dynamics of p53 by using single molecule microscopy. In addition, we are developing a new strategy of protein design.Observation of protein folding dynamics using the single-molecule fluorescence tracking system developed in this group
Representative recent publications
- Kiyoto Kamagata, Saori Kanbayashi, Masaya Honda, Yuji Itoh, Hiroto Takahashi, Tomoshi Kameda, Fumi Nagatsugi, Satoshi Takahashi, “Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains”, Sci. Rep. 10, 580 (2020)
- Kiyoto Kamagata, Eriko Mano, Yuji Itoh,, Takuro Wakamoto, Ryo Kitahara, Saori Kanbayashi, Hiroto Takahashi, Agato Murata, Tomoshi Kameda, “Rational design using sequence information only produces a peptide that binds to the intrinsically disordered region of p53”, Sci. Rep. 9, 8584 (2019)
- Itoh Y.,Murata A., Takahashi S., Kamagata K., “Intrinsically disordered domain of tumor suppressor p53 facilitates target search by ultrafast transfer between different DNA strands”, Nucleic Acid Res. 46, 7261-7269 (2018)
- Subekti D. R. G., Murata A., Itoh Y., Fukuchi S., Takahashi H., Kanbayashi S., Takahashi S., Kamagata K., “Disordered linker in p53 participates in non-specific binding to and 1D sliding along DNA revealed by single-molecule fluorescence measurements”, Biochemistry 56, 4134-4144 (2017)
- Kiyoto Kamagata, Agato Murata, Yuji Itoh, Satoshi Takahashi, “Characterization of facilitated diffusion of tumor suppressor p53 along DNA using single-molecule fluorescence imaging”, J. Photochem. Photobiol. C photocem. Rev. 30, 36-50(2017)
- Igarashi C.,Murata A.,Itoh Y.,Subekti D.R.G.,Takahashi S.,Kamagata K., “DNA garden: A simple method for producing arrays of stretchable DNA for single-molecule fluorescence imaging of DNA binding proteins”, Bull. Chem. Soc. Jpn. 90, 34-43(2017)
- Masataka Saito, Supawich Kamonprasertsuk, Satomi Suzuki, Kei Nanatani, Hiroyuki Oikawa, Keiichiro Kushiro, Madoka Takai, Po-Ting Chen, Eric Hsin-Liang Chen, Rita Pei-Yeh Chen, and Satoshi Takahashi, “Significant Heterogeneity and Slow Dynamics of the Unfolded Ubiquitin Detected by Confocal Method of Single-Molecule Fluorescence Spectroscopy” J. Phys. Chem. B 120, 8818-8829, (2016)
- Yuji Itoh, Agato Murata, Seiji Sakamoto, Kei Nanatani, Takehiko Wada, Satoshi Takahashi, Kiyoto Kamagata, “Activation of p53 Facilitates the Target Search in DNA by Enhancing the Target Recognition Probability”, J. Mol. Biol. 428, 2916–2930,(2016)
- Hiroyuki Oikawa, Yuta Suzuki, Masataka Saito, Kiyoto Kamagata, Munehiro Arai and Satoshi Takahashi "Microsecond dynamics of an unfolded protein by a line confocal tracking of single molecule fluorescence" Sci. Rep. 3, 2151 (2013)
- Kamagata K, Kawaguchi T, Iwahashi Y, Baba A, Fujimoto K, Komatsuzaki T, Sambongi Y, Goto Y, Takahashi S "Long-Term Observation of Fluorescence of Free SingleMolecules To Explore Protein-Folding Energy Landscapes" J. Am. Chem. Soc. 134, 11525-11532 (2012)
- Tsuyoshi Konuma, Tetsunari Kimura, Shuzo Matsumoto, Yuji Goto, Tetsuro Fujisawa, Alan R.Fersht and Satoshi Takahashi "Time-Resolved Small-Angle X-ray Scattering Study of the Folding Dynamics of Barnase" J. Mol. Biol. 405,1284-1294 (2011)